Public Health
Viral Surveillance
Helix partners with organizations like the CDC to monitor emerging SARS-CoV-2 variants. We've played an integral role in identifying Alpha, Delta and Omicron across the U.S.
Resources
During the COVID-19 pandemic, Helix scaled overnight to support health systems and public partners
150,000
daily COVID-19 test capacity with next day turnaround time
Hundreds of thousands
of COVID-19 viral sequences analyzed and reported
First private sector partner
to identify 51 out of the first 54 cases of the Alpha variant in the US and predict that Delta cases would make up at least half of all COVID-19 infections by mid-summer 2021
Frequently Asked Questions
Helix is working together with the CDC and other public health agencies on national viral surveillance efforts. Helix receives nasal swabs as part of our normal workflow for COVID-19 testing as the lab processing samples for several commercial partners. After processing these samples and reporting SARS-CoV-2 detection, positive samples may be selected for follow-on sequencing, based on demographics, sample quality, test characteristics, or simply random selection. These samples are submitted for sequencing where they are further processed and aligned to the SARS-CoV-2 reference genome (NC_045512.2) to obtain sequencing results. Raw sequence information is transferred to Helix as well as to the CDC for nationwide testing and surveillance. Helix and/or our partners may also submit raw data, consensus sequence, and metadata to NCBI and GISAID. Helix monitors the data that becomes publicly available and, in turn, makes these identifiers available in an aggregated report in our public Github repository: https://github.com/myhelix/helix-covid19db.
Helix further processes the sequence data to identify phylogenetic lineage and clade using Pangolin and Nextclade. These annotations are also available in our GitHub, which is the data that backs our Dashboard. For Variants of Interest or Concern (VOI, VOC) such as Delta, we further notify State Departments of Health for contact tracing within 24 hours of receipt of sequencing results.
We use the residual samples after completion of processing the Helix® COVID-19 Test or other COVID-19 tests as requested, which uses samples collected from anterior nares swabs in saline. The assay is a 3-gene target panel based on Thermo Fisher Scientific's TaqPath™ COVID-19 Combo Kit. You can read about our testing program here: helix.com/covid19. In order to qualify for sequencing, we require sufficient concentration of virus in the original sample. Therefore we have set a threshold of Cq(N gene) < 27, and a minimum volume of 500ul/sample.
We use widespread random sampling in order to detect other variants of concern and monitor the rise of new variants. We continue to work with the CDC to update our sampling strategy based on their feedback and public health needs.
Not all residual samples have enough volume or enough virus particles to have success during sequencing. Regardless of sampling strategy, we filter all samples for Cq (N gene or ORF1ab) < 27 prior to sequencing to ensure robust sequencing efforts.
It typically takes 1-2 weeks to sequence selected samples. As the program continues to grow, we expect the turnaround time to decrease - there are currently 2-3 sequencing runs per week. Note that this timeframe is for viral sequencing. General COVID-19 testing to detect SARS-CoV-2 usually takes less than 24 hours after receipt in the Helix laboratory.
- The Helix COVID-19 biobank and viral surveillance program has been reviewed and approved by Western Institutional Review Board Protocol WIRB#20203438.
- We are currently working with the CDC, San Diego County, and some additional partners who request individual samples be sequenced.
- Patient privacy is our utmost concern. Within our research protocol, we have received a waiver of consent for a limited dataset under HIPAA regulations for the purposes of public health (Privacy Rule (45 CFR § 164.512(b)). The Helix research team receives no individually identifiable information beyond the limited dataset that includes sample collection date and zip code. This information is sensitive and is not revealed in the data available for public download in GitHub.
Yes, confirmation of any VOC can only be made with viral sequencing. Viral sequencing with Helix will identify not just the variants that have already been identified as VOCs, including Alpha (B.1.1.7) and Delta (B.1.617.2), but the results would enable identification and tracking of any novel variants that arise. Helix performs regular reprocessing of previously sequenced viral genomes to ensure retrospective annotation of newly classified variants.
There are numerous steps that happen between a sample being sequenced and the data appearing on GISAID. After a sample is sequenced, we have to analyze and assemble the data. After that, the sequences are submitted to GISAID, which sometimes requires some troubleshooting. There is typically a 2 ½ week lag from collection of the initial sample to upload of the data on GISAID, but may be longer for some samples.
Sequencing data generated from samples originating at Helix can be obtained from two public repositories: GISAID and NCBI. Deposition into these public repositories is being done by our collaborators at CDC and Scripps.
All samples that originate at Helix should have “STM” in the sample name. This string can be used when searching for samples at NCBI or GISAID.
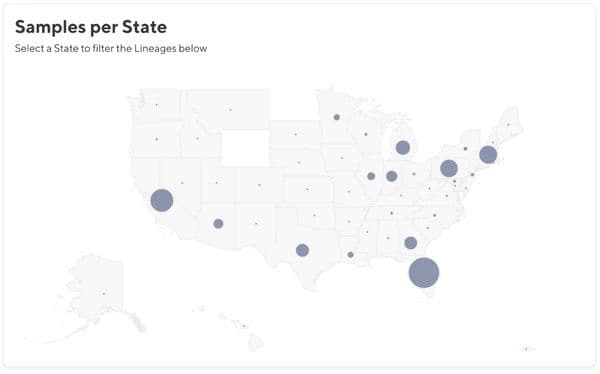
In order to assist with your research, you can find a summary of the sequence data at our public GitHub repository here: https://github.com/myhelix/helix-covid19db
We make limited metadata including State and NCBI accession number for Helix samples available on our public GitHub repository, which can be used to filter for your state of interest: https://github.com/myhelix/helix-covid19db
- For press inquiries, please contact press@helix.com.
- For establishing new viral surveillance programs with Helix, please contact covid@helix.com.
- For public health reporting, please contact dphreporting@helix.com.
For research questions, please contact research@helix.com. - Follow @my_helix on Twitter where we will update about the program and significant research findings.
Get in touch
To learn more about our COVID-19 response & viral surveillance, complete this form and we’ll be in touch soon.
If you have questions about your COVID-19 test result, please email support@helix.com.