Update on the Helix, Illumina surveillance program: B.1.1.7 variant of SARS-CoV-2, first identified in the UK, spreads further into the US

Most recent update: January 11th, 2021
What’s new?
- Helix has identified 74 cases of B.1.1.7 (up from the 51 reported in our January 6 update).
- B.1.1.7 appears to currently represent 0.27% of all positive tests and 43% of SGTF samples, up from 0.17% and 34% last week, respectively.
- B.1.1.7 has been found in 3 additional states (TX, MN, IN), adding to the 4 previously reported (CA, FL, PA, GA)
- B.1.1.7 has been found in 4 of the 5 states where we have statistical power to detect it (CA, FL, PA, IN), as well as 3 additional states where we are underpowered (GA, TX, MN).
- We’ve added a breakdown of statistics for the three states in which we have sequenced the highest numbers of samples
- Despite having the power to detect it, we have yet to identify B.1.1.7 in MA
- 43% of total SGTF sequenced samples are identified as the B.1 variant, which was initially associated with the spread of COVID-19 in Italy during the early months of 2020 but has not been found to be more contagious
- Presence of the B.1 variant varies significantly by state, however, representing 80% of SGTF samples in MA, 56% of samples in FL, and 2% of samples in CA.
Our current understanding of B.1.1.7 in the United States
The SARS-CoV-2 variant (B.1.1.7) strain—first identified in the United Kingdom—has garnered much of the world’s attention in recent weeks. This variant is known to carry several mutations that culminate in a more infectious strain of the virus and presents a significant threat to public health.
We first reported the potential spread of the B.1.1.7 variant in the United States based on the occurrence of S gene dropout—a phenomenon where qRT-PCR testing fails to detect the presence of the virus’ S gene, owing to a deletion mutation affecting amino acids H69 and V70 (this is also known as S gene target failure, or SGTF). This deletion is one of several mutations that distinguish the B.1.1.7 from other SARS-CoV-2 strains.
Subsequent viral sequence analysis of SGTF samples using Illumina’s COVIDSeq Test confirmed that only a subset of these samples harbored all variants that define B.1.1.7. The remaining SGTF samples were missing the other mutations that are characteristic of B.1.1.7. We have continued to sequence SGTF samples, which has led to the identification of a total of 74 individuals with the B.1.1.7 variant strain in multiple states. We have assigned each of the SGTF sequences to a phylogenetic clade in order to understand the recent evolution of this virus (results shown below).
One strain that stood out in this week’s data is the B.1 variant which appears to make up a large proportion of SGTF samples (43%). B.1 is a variant of the SARS-CoV-2 virus that was initially associated with the spread of COVID-19 in Italy during the early months of 2020. This variant of the virus is similar to B.1.1.7 in that it harbors the D614G missense mutation and, in some populations, may also carry the H69del mutation (eliciting SGTF). However, B.1 is lacking the full mutational profile that defines B.1.1.7 and is not more contagious than earlier strains of SARS-CoV-2.
It is noteworthy that we do not see B.1.1.7 in MA, despite being well powered to detect it based on the number of samples we have sequenced (99.9% power to detect the mean nationwide rate of 40%; 80% power to detect if at least 5% of SGTF samples were caused by B.1.1.7). We have, however, noticed that 80% of sequenced SGTF samples from MA are identified as having the B.1 variant. By comparison, B.1 is identified in 56% of sequenced SGTF samples in Florida and only 2% of samples in CA. The significance, if any, of B.1 overrepresentation in MA is not yet known.
To monitor trends in B.1.1.7 transmission, as well as the dynamics of other strains harboring the H69/V70 deletion, we will continue to regularly sequence high-quality SGTF samples with N gene Cq<28 from our lab and place these strains on the SARS-CoV-2 phylogeny. While growing in prevalence, SGTF samples represent a small subset of all positive Helix® COVID-19 Tests. We also plan to expand this surveillance program past SGTF samples to proactively monitor for the emergence and flow of new strains.
Please check back frequently: We will be updating the figures below as our data continues to evolve.
This work is being done with the support of the CDC and in partnership with Illumina. Sequences will be deposited regularly to GISAID and Genbank. As a matter of public health, all B.1.1.7 strains are also reported to relevant public health departments for contact tracing efforts.
Current stats based on Helix data
At a glance
| Metric | Result |
|---|---|
| Total number of SGTF samples sequenced | 187 |
| Total number of B.1.1.7 cases detected | 74 |
| Total states with B.1.1.7 to date (CA, FL, PA, IN, GA*, TX*, MN*) | 7 |
| Total states with more than 5 SGTF samples sequenced (95% power to detect at least 1 B.1.1.7 given the overall mean—among sampled states—of 40%) | 5 |
*B.1.1.7 detected in these states despite being underpowered to detect it (having sequenced fewer than 5 SGTF samples).
Longitudinal trends in SGTF and B.1.1.7 data
National data ▼
| Dec 13 – 19 | Dec 20 – 26 | Dec 27 – Jan 2 | |
|---|---|---|---|
| Percent of positives that are SGTF | 0.5% | 0.5% | 0.6% |
| Percent of SGTF sequenced samples that are B.1.1.7 | 10.0% | 34.2% | 43.2% |
| Percent of positive tests that are B.1.1.7 (extrapolated) | 0.1% | 0.2% | 0.3% |
The three states where we have done the most sequencing
California ▼
| Dec 13 – 19 | Dec 20 – 26 | Dec 27 – Jan 2 | |
|---|---|---|---|
| Percent of positives that are SGTF | 0.4% | 0.5% | 0.5% |
| Percent of SGTF sequenced samples that are B.1.1.7 | NA | 72.7% | 71.4% |
| Percent of positive tests that are B.1.1.7 (extrapolated) | NA | 0.3% | 0.4% |
Florida ▼
| Dec 13 – 19 | Dec 20 – 26 | Dec 27 – Jan 2 | |
|---|---|---|---|
| Percent of positives that are SGTF | 1.3% | 1.2% | 1.6% |
| Percent of SGTF sequenced samples that are B.1.1.7 | 20.0% | 41.7% | 42.3% |
| Percent of positive tests that are B.1.1.7 (extrapolated) | 0.3% | 0.5% | 0.7% |
Massachusetts ▼
| Dec 13 – 19 | Dec 20 – 26 | Dec 27 – Jan 2 | |
|---|---|---|---|
| Percent of positives that are SGTF | 2.9% | 3.2% | 4.0% |
| Percent of SGTF sequenced samples that are B.1.1.7 | 0.0% | 0.0% | 0.0% |
| Percent of positive tests that are B.1.1.7 (extrapolated) | 0.0% | 0.0% | 0.0% |
Data visualization
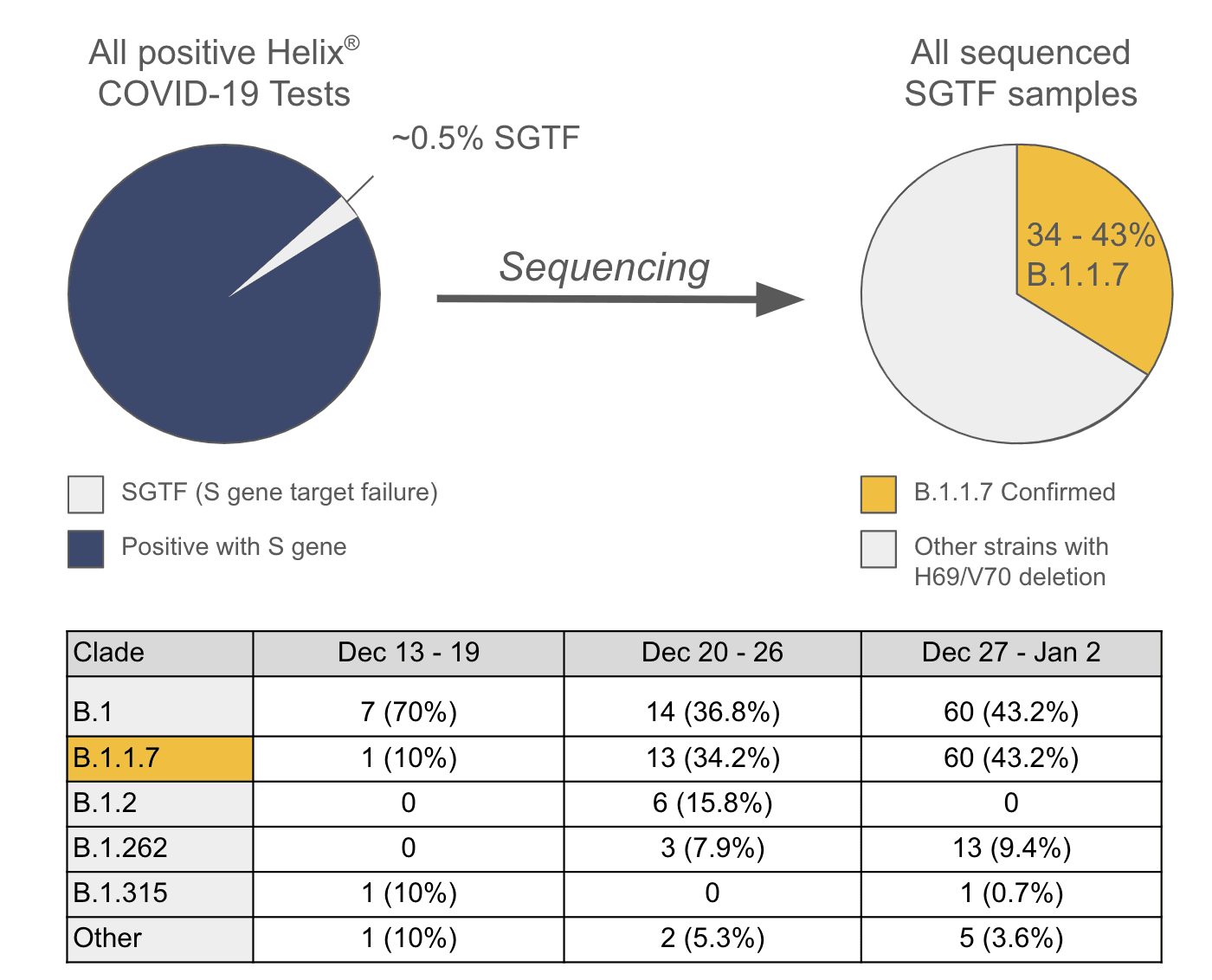
Figure 1. Analysis pipeline and SARS-CoV-2 Clades in SGTF samples Breakdown of the analysis pipeline used to identify B.1.1.7 samples from all positive Helix COVID-19 Test samples to date. “Other” represents all clades that were seen only once.**Updated 01/11/2020
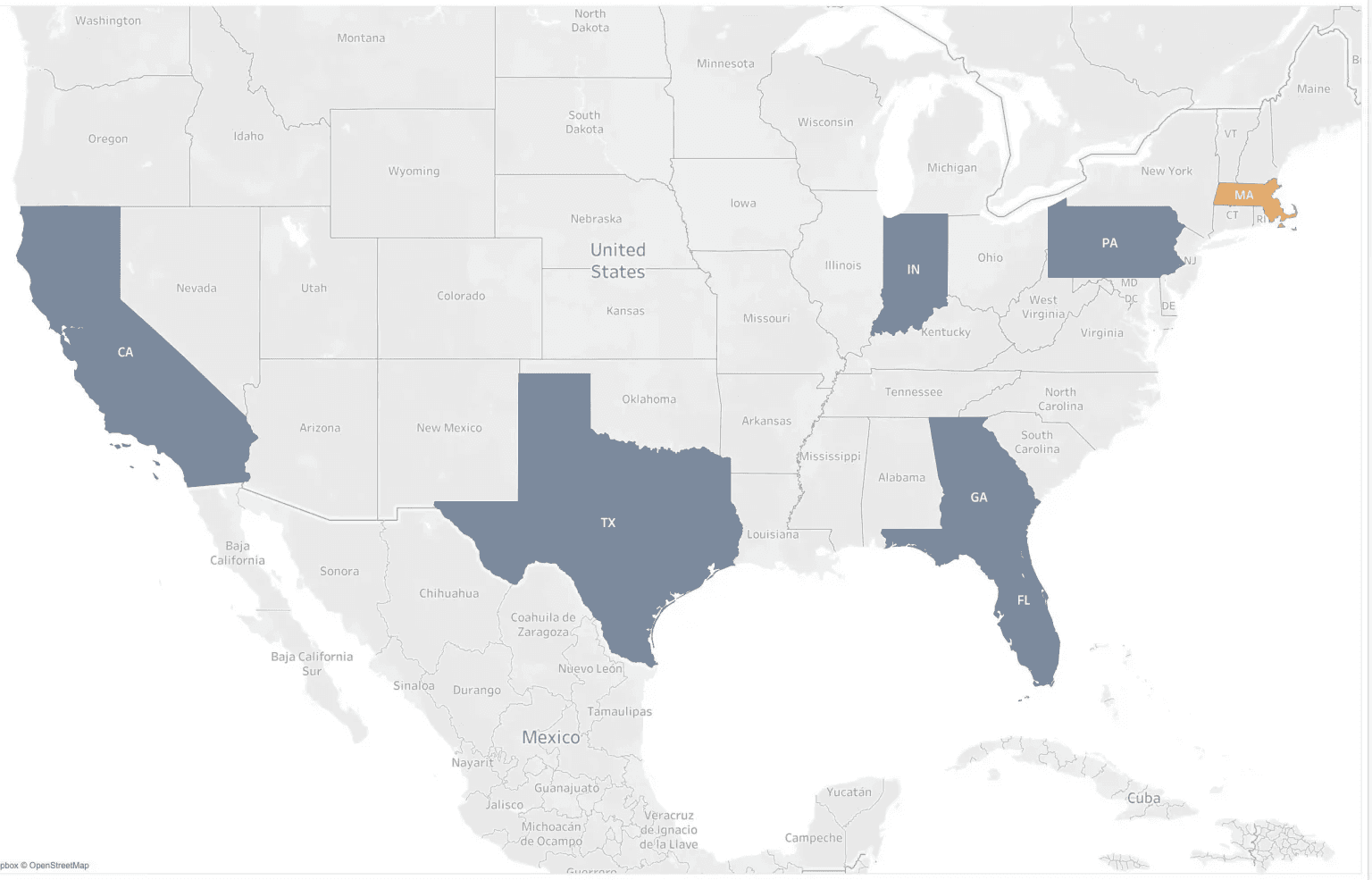
Figure 2. Distribution of the B.1.1.7 variant in the US. Shown are states with at least 1 B.1.1.7 case found in Helix sequence data (blue), states with 0 B.1.1.7 but more than 5 sequenced SGTF samples (orange, 95% power to identify B.1.1.7 given mean percentage), and states with <=5 SGTF sequenced (gray, not yet powered for discovery).
Categories