My DNA story: Dr. Eric Schadt, founder and CEO of Sema4

April 25th is DNA Day! In celebration, we’ve worked with some of our partners to hear their DNA stories—how they got into genetics, what inspires them most, and what excites them about the future of personal genomics.
Eric Schadt, Ph.D., is founder and CEO of Helix partner Sema4, a health information company that is creating practical tools to help patients, clinicians, and researchers better diagnose, treat, and prevent disease. Dr. Schadt also serves as Dean for Precision Medicine at the Icahn School of Medicine at Mount Sinai. He was previously Founding Director of the Icahn Institute for Genomics and Multiscale Biology, and Professor and Chair of the Department of Genetics and Genomic Sciences. Dr. Schadt is an expert on constructing predictive models of disease that link molecular biology to physiology to enable clinical medicine. Over the past 20 years, he has built groups and companies (Merck, Rosetta, Sage Bionetworks, Pacific Biosciences, Icahn Institute, and now Sema4) to elucidate the complexity of human diseases. He has published more than 350 peer-reviewed papers in leading scientific journals, and contributed to a number of discoveries relating to the genetic basis of common human diseases such as diabetes, obesity, and Alzheimer’s disease.
My DNA journey was a rather long and winding one and involved scaling a few mountains along the way. My academic training began in computer science and math as an undergraduate and then, as a graduate student, I started traversing an intellectual wilderness to explore how far I could push myself in what I considered to be the most intellectually challenging pursuit I could manage: pure mathematics. But as I progressed, coming to grips with how smart I was and how smart I was not, I began thinking more about my own life course, my purpose, and our more collective purpose. I questioned everything,pondered the existence of God, and how our world came into being. With a more scientific and curious mind, it didn’t take long for me to come face to face with evolution, and I arrived at this place at a point in time (1991) when molecular biology had come to dominate the life sciences, and we were at the dawn of the genomics revolution.
I began attending biology seminars, where I learned about the Human Genome Project and the growing use of molecular profiling techniques, including DNA genotyping and sequencing technologies, to systematically characterize phylogenetic relationships among species. I was fascinated by the idea that changes in DNA affecting the activities of proteins highly conserved across diverse species could be simultaneously assayed and compared, allowing organisms to be ordered with respect to one another, and the evolutionary pressures acting on those changes inferred from these relationships. But, as I dug deeper into molecular biology to better understand the mechanisms that enabled living systems to evolve and adapt, the simplicity of the models proposed to explain complex biological processes seemed grossly oversimplified, from the central dogma of biology (DNA leading to RNA leading to proteins) to the linearly ordered pathways used to explain core molecular processes defining life. How could the millions of molecular species making up a cell, the complex web of cellular interactions and communications comprising a tissue (including the billions of neurons in the human brain and the many trillions of connections between the neurons), the coordinated activity of many organs giving rise to coherent living beings, and the emergent behaviors such as consciousness arising from such complex systems, all be explained by simple linear pathways?
I began to realize that I had landed at a critical crossroads in the life sciences, where biology was on the verge of being transformed from the study of single genes or proteins, one or a handful at a time, to many or all genes in a genome simultaneously and across populations of individuals. I could see that the qualitative nature of the then-current biology was going to give way to a more quantitative version that considered hundreds of thousands to millions of variables at a time and could result in the generation of nearly endless numbers of hypotheses for testing. It became apparent to me that the only way to be successful in that arena of big data biology, the only way to make progress, would be by employing advanced computational algorithms to sift through the vast oceans of data, in order to prioritize hypotheses for experimental pursuit and to produce the frameworks necessary for representing knowledge and understanding in a dynamic, adaptive, and evolving manner.

That was the moment I set my more purposeful genomics trajectory
That was the moment I set my more purposeful genomics trajectory. I hypothesized that the life and biomedical sciences would need quantitative guys like me to effectively ride the technology wave rapidly coming their way. Pioneers in the DNA sciences such as Lee Hood, who was defining his own systems biology vision just as I began to stumble upon my own, served as sources of inspiration and validation that I was bouncing onto the right trajectory at the exact right time. So, after getting to PhD candidacy, I abandoned my pure math pursuits and entered a new chapter in my academic journey focused on biomathematics, simultaneously studying biology (molecular genetics in my case) and applied mathematics in the hope of playing a role in defining how to approach biological systems in a more quantitative, higher dimensional way.
While a great many technologies in the 1990s and early 2000s began to empower biology to move at Moore’s Law speed, one technology in particular—next-generation nucleic acid sequencing—was moving fastest of all, enabling biology to not only participate in the era of big data, but also to be a central contributor to it. Whereas semiconductor-based technologies move at Moore’s Law speed, next-generation sequencing has been progressing at super Moore’s Law speed—the only technology in the history of humankind that has moved with such speed, as far as I am aware. I leaped without looking and without much thought onto this wave, leading the
computational genomics group that provided the first functional annotation of the initial draft of the human genome by the Human Genome Project in 2001 (Nature, 2001), then leading the first group to systematically and quantitatively characterize the relationship between variations in DNA and gene expression (which led to the first “eQTL” studies in plants and mammals, including humans—Nature, 2003). We were also the first to comprehensively leverage DNA variation as a systematic source of perturbation to causally infer relationships among molecular traits (such as gene expression, metabolomics, and proteomics) and between molecular and clinical traits, moving for the first time beyond correlation-based methods that were dominating the high-dimensional biology space (Nature, 2003 and Nature Genetics, 2005). Causal reasoning approaches, in collaboration with others such as my long-time colleague Jun Zhu, quickly led to network-based approaches that were capable of inferring causal relationships among many thousands of traits (molecular, clinical, and physiologic), providing a structure-based learning framework that could be used to uncover new knowledge and understanding in an objective, data-driven way.
As a result of these advances made by my groups and collaborators, in addition to many other groups, I have dedicated myself to integrating large, diverse sets of data to construct causal models of disease, drug response, and other interesting phenotypes such as wellness. These pursuits have resulted in the identification and validation of novel disease genes, disease networks, optimal points for therapeutic intervention, and biomarkers for patient stratification. The efforts I have led at the Icahn School of Medicine at Mount Sinai in New York have leveraged the rapidly accumulating base of genomic knowledge and translated it into clinical care, encompassing genomic tests focused on reproductive health and oncology, and the characterization of somatic genomes for more accurate diagnosis and treatment decisions in cancer patients.
However, as I immersed myself into one of the largest health systems in the country, the Mount Sinai Health System, it became painfully obvious that we simply did not have access to the scale of data needed to derive the most meaningful insights and build the most predictive models. Given enough high-dimensional, longitudinal data—in the realm of many millions of individuals—these models would clinically impact our ability to map out individualized health course trajectories that would, in turn, optimize our ability to diagnose, treat, and prevent disease in patients. DNA is an important dimension for mapping out these individualized health course trajectories, but other dimensions of data at the molecular, physiologic, and micro-/macro-environmental levels are also essential to form a holistic view of a patient over time.
I founded Sema4 to enable us to obtain the scale of data needed to generate clinically actionable information capable of guiding day-to-day decision-making around one’s health. We want Sema4 to be among the first to achieve this type of scale in the health arena, making possible a new vision for information-rich health care—a vision that puts consumers and patients in control. We are a patient-centered predictive health company, with a broader mission than being just a leading diagnostic testing company. The testing component of Sema4 is our growth-hack engine for getting to the numbers and scale of data we need to fundamentally transform our ability to define and put into practice individualized health course trajectories. Ultimately, we hope to build a vast information store and biobank in partnership with patients that will change our understanding of human disease.
We want to empower individuals with their healthcare information
At the same time, we want to empower individuals with their healthcare information. Our goal is to help our patients manage their health information, to aggregate it for them in useful ways, and enable them to gain insights from it and to share their information with whomever they want. By leveraging our diagnostic testing capabilities to partner with the individuals we are testing, we can convey more meaningful results and insights that in turn help better guide them along their health course journeys, as well as engage them on an ongoing basis. To get a full picture of a person’s health and wellbeing, we need much more than just a DNA sample and medical records. Therefore, we are engaging with patients to get ongoing data about their health, their activity levels, their diet, their environment, and so on. I believe the best way to promote wellness is to understand living things as vast information networks.
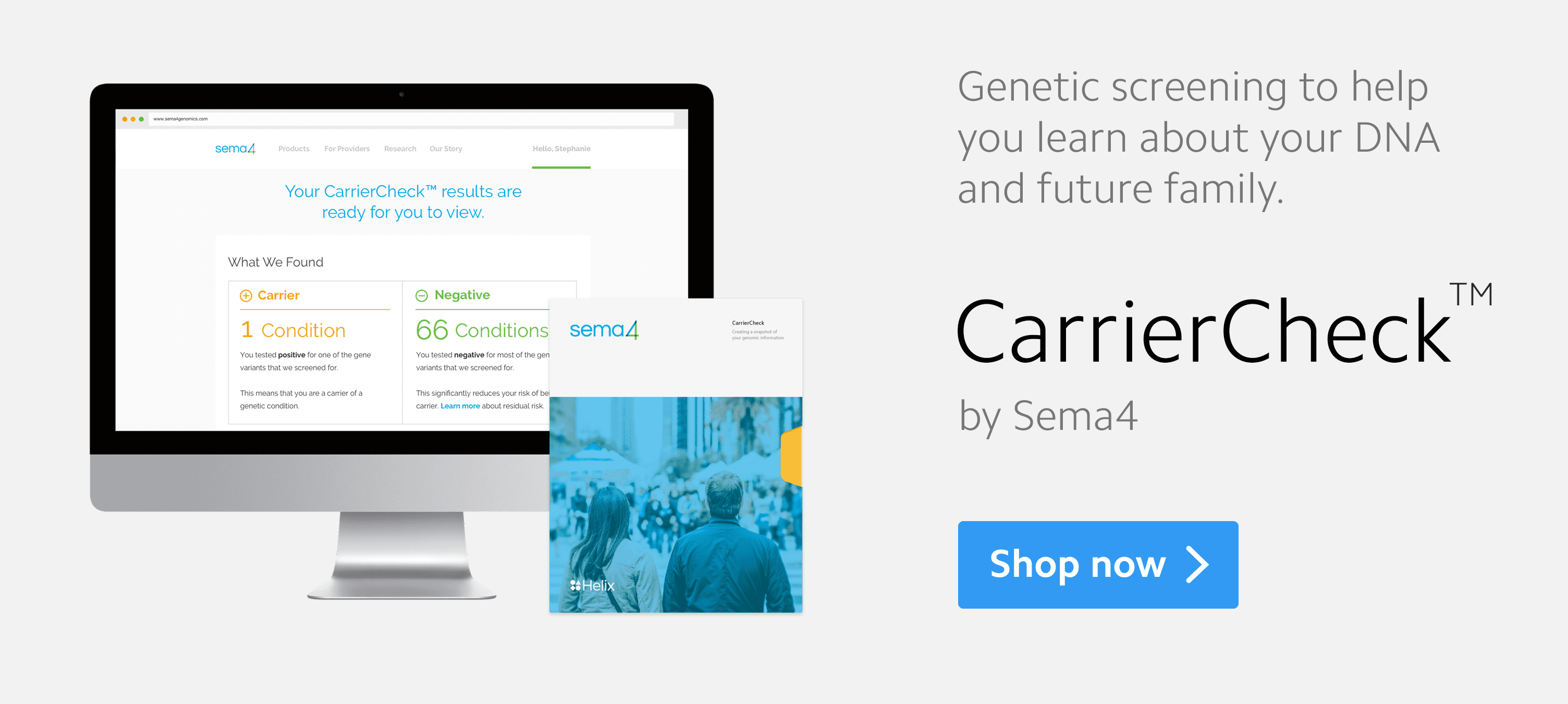
Helix shares in many aspects of our ethos—forming meaningful, ongoing relationships with consumers, empowering them with data built up around them, empowering researchers or others who require access to larger-scale data to extract meaningful insights that they can feedback to the consumer, and democratizing access to DNA data—and so they were an obvious partner for the launch of CarrierCheck, our 67-gene carrier screen available through the Helix platform. Our other current tests from Sema4 include expanded carrier screening, supplemental newborn screening, pharmacogenetic testing for children and adults, heritable cancer screening, and targeted oncology testing in somatic genomes, in addition to a diversity of more targeted tests to help aid in the diagnosis of specific conditions such as autism. In the next few months, we will be launching a number of additional diagnostic tests, including more comprehensive oncology tests, and broadening our reproductive health portfolio with non-invasive prenatal testing, pre-implantation genetic diagnosis/screening, as well as substantial enhancements to our carrier screening. And we will, of course, be working towards our goal of collating data on more than 10 million participants, both through our own testing and through partnerships with large health systems and laboratory testing companies.
The next step in personal genomics will be whole-exome sequencing, which Helix is already enabling today. Affordable, rapid whole-genome sequencing will follow hot on its heels. We are on the cusp of a very exciting phase in DNA science. Now is a fantastic time for young people to be getting into genetics, not just for those interested in biology, but also for those—like me—who have a head for numbers and computing. And for those already training in another discipline, I think my DNA journey shows that you can always change your path and apply your skills to a new field.